LAB REPORT
Science and Technology Making Headlines
March 1, 2019


Melting sea ice is just one of the effects of global climate change.
Three is the key
Three key events in the history of climate science hit their 40th anniversary this year and Lawrence Livermore scientists say they make up the “gold standard” level of certainty of man-made global warming.
During the same decade that saw the Beatles break up, the Watergate scandal, the first Earth Day and the opening of the movie Star Wars, efforts to identify human-caused climate change were in their infancy.
Three key events in the 1970s heralded the end of that infancy. The events were the release of an influential National Academy of Sciences report, the publication of a key paper on anthropogenic signal detection and the start of satellite temperature measurements. All three events occurred in 1979. The research appears in Nature Climate Change.
“We can’t afford to ignore such clear signals,” said Stephen Po-Chedley, an LLNL co-author who has developed temperature records from satelllite data.


The Deccan volcanic stratigraphy, manifest in massive mountains. They are spread over hundreds of thousands of square kilometers, with individual layers of solidified lava than can be connected over enormous distances.
Volcanoes vs. T-Rex: Who wins?
As it turns out, the volcanoes and a giant asteroid actually won and the dinosaurs died off.
Solidified lava from ancient volcanoes, over a mile thick, covers a portion of west-central India called the Deccan Traps. Contained in the striped rock are secrets a Lawrence Livermore scientist and collaborators are only beginning to uncover — secrets that could partially rewrite the story of the dinosaur mass extinction 66 million years ago.
LLNL’s Kyle Sampeton’s team exhibits the most precise Deccan eruptive timeline yet, aligning the dinosaur extinction with massive volcanic events that occurred around the same time of the Chicxulub meteorite impact in Mexico. The meteorite impact has long been viewed as the primary (if not sole) cause of the mass extinction at the Cretaceous-Paleogene boundary, the geological moment when many creatures from the age of dinosaurs vanish from the fossil record.
However, the team’s new findings suggest Deccan eruptions were highly nonlinear, with four major volcanic pulses. According to their eruptive age model, the largest of these volcanic pulses immediately precedes the mass extinction event with an approximate 90 percent probability.


New research suggests snowpack loss in the Sierra Nevada will fast-track in the coming decades.
Sierra snowpack melt is steady, but not for long
Snowpack loss in the northern Sierra Nevada will likely accelerate in the coming decades.
Scientists at Lawrence Livermore National Laboratory (LLNL), Oregon State University and the University of Washington found that natural swings in the sea surface temperature over the Pacific Ocean have resulted in atmospheric circulation changes that have muted the effect of climate change on snow in the Sierras since the 1980s.
Although the amount of water contained in the snowpack has declined over the past century, it has been surprisingly stable since the 1980s, despite 1 degree Celsius of warming over the same period.
“A comparison between temperature and snowpack changes suggests that snowpack is quite resilient to warming,” said LLNL scientist Stephen Po-Chedley. “However, we found that the contribution of global warming to Western U.S. snowpack loss has in reality been large and widespread since the 1980s, but mostly offset by natural variability in the climate system.”
The result points to a faster rate of snowpack loss in coming decades, when the impact of global warming is more likely to be amplified, rather than offset, by natural variability.

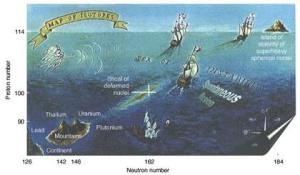
The fabled island of stability should be at coordinates where there is a “magic” number of protons and neutrons.
Pushing the boundaries one atom at a time
Scientists are hoping to stretch the periodic table even further, beyond tennessine (element 117) and three other recently-discovered elements (113, 115 and 118) that completed the table’s seventh row. Producing the next elements will require finessing new techniques using ultrapowerful beams of ions.
But questions circulating around the periodic table’s limits are too tantalizing not to make the effort. It’s been 150 years since Russian chemist Dmitrii Mendeleev created his periodic table. Yet scientists still can’t answer the question: Which is the heaviest element that can exist?
At the far edge of the periodic table, elements decay within instants of their formation, offering very little time to study their properties. Scientists keep pushing these superheavy elements further as part of the search for what’s poetically known as the island of stability.
Driving all this effort is a deep curiosity about how elements act at the boundaries of the periodic table. “This might sound corny, but it’s really just [about] pure scientific understanding,” said nuclear chemist Dawn Shaughnessy of Lawrence Livermore National Laboratory. “We have these things that are really at the extremes of matter and we don’t understand right now how they behave.”


Lawrence Livermore scientists and engineers are combining mechanical computing with 3D printing as part of an effort to create “sentient” materials that can respond to changes in their surroundings, even in extreme environments. Pictured, from left, are LLNL researchers Julie Jackson Mancini, Logan Bekker, Andy Pascall and Robert Panas. Photo by Julie Russell/LLNL
It’s only logical
Engineers and scientists from Lawrence Livermore and UCLA are 3D printing mechanical logic gates, the basic building blocks of computers that can perform any kind of math calculations.
These 3D-printed logic gates could be used to build just about anything, researchers said, embedded into any type of architected material and programmed to react to its environment by physically changing shape without the need for electricity. This would make them useful in areas of high radiation, heat or pressure.
“If you embedded logic gates into material, that material could sense something about its environment,” says lead researcher LLNL Andy Pascall. “It’s a way of having a responsive material — we like to call it a sentient material — that could have complicated responses to temperature and pressure.”
Mechanical logic gates, while are not as powerful as typical computers, could prove useful in rovers sent to hostile environments such as Venus, or else in low-power computers intended to survive nuclear or electromagnetic pulse blasts that would destroy electronic devices, researchers said.





