New research shows successful ab initio crystal structure prediction of energetic materials
 (Download Image)
(Download Image)
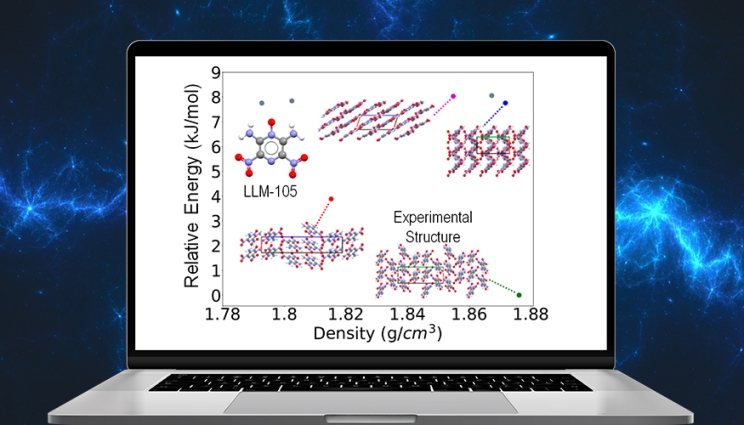
The figure shows the energy ranking, where lower energy means higher stability, as a function of density for the most stable crystal structures generated by the algorithm. The experimentally observed structure of LLM-105 is the most stable and has the highest density. The algorithm also produces putative crystal structures in a variety of packing motifs. Crystal structures in which the molecules are arranged in layers have been associated with low sensitivity, and the structure indicated by the purple marker has a layered packing motif. This demonstrates how simulations can help discover new crystal structures with desirable properties.
New research by Lawrence Livermore National Laboratory researchers and collaborators from Carnegie Mellon University (CMU) demonstrates that crystal structure prediction is a useful tool for studying the various ways the molecules can pack together, also known as ubiquitous polymorphism, in energetic materials. The research also shows promise of becoming an integral part of the energetic materials development pipeline.
Experimental synthesis of novel energetic materials can be a costly, time-consuming and hazardous process, sometimes requiring a significant amount of trial and error. If material properties could be predicted ahead of time, compound selection and solid form development process could be streamlined.
Predicting materials properties with a physics-based approach requires knowledge of the crystal structure. The work, featured in the Aug. 16 edition of Crystal Growth & Design by the American Chemical Society, has demonstrated that the crystal structure of commonly used energetic materials LLM-105, RDX, and HMX can be predicted using ab initio (i.e. without experimental input) crystal structure prediction methods, showing the viability and usefulness of this approach.
The work was led by Brad Steele and Anna Hiszpanski from LLNL’s Materials Science Division in collaboration with Noa Marom from the Materials Science & Engineering Department at CMU. The work was funded by the Lab’s Laboratory Directed Research and Development (LDRD) program, with T. Yong-Jin Han serving as primary investigator. CMU Ph.D. student, Dana O’Connor, received a DOE Office of Science Graduate Student Research (SCGSR) award to conduct research at LLNL.
Steele compared crystal structure prediction to going on a long road trip and trying to pack as much stuff as possible in your car’s trunk, rearranging the stuff until the space is optimally packed.
“In our case, the ‘stuff’ are molecules, and the trunk is the unit cell — which is a ‘box’ with variable shape and size — that is repeated periodically in three dimensions to form the crystal lattice,” Steele said. “The way molecules are packed in the crystal affects key properties and performance parameters of energetic materials, such as the density and the shock sensitivity.”
Steele said the paper describes the workflow of the crystal structure prediction simulations. The algorithm is a physics-based approach that works by randomly generating crystal structures, optimizing them and ranking the resulting structures by their predicted stability.
First, a machine learned model trained on experimental data from the Cambridge Structural Database estimates the unit cell volume (i.e. the size of the box in which the molecules will be packed) based on the molecule’s structure. Second, the random structure generator, Genarris, produces an initial set of putative crystal structures by generating unit cells with a distribution around the target volume and placing molecules inside them in all the ways allowed by space group symmetries, such that no two molecules are too close to each other. Third, the set of structures generated by Genarris are used as the initial population for the genetic algorithm (GA), GAtor. A GA performs optimization by mimicking evolution. Based on survival of the fittest, parent structures are selected for “mating” and their “genes” (e.g., the size and shape of the unit cell, the molecules’ positions and orientations, and space group symmetries) are combined (crossed-over) or altered (mutated) to form offspring. This is repeated in order to propagate the structural features associated with high stability until the optimal crystal structure is found.
“The work impacts the mission of the Lab because it is a new method that could be useful in the development of novel energetic materials,” said Brad Steele. “The paper demonstrates that if we have knowledge of the molecular structure of an energetic material, then we can predict its crystal structure and then a variety of other important material properties.”
Contact
 Michael Padilla
Michael Padilla
[email protected]
(925) 341-8692
Tags
Advanced Materials and ManufacturingMaterials Science
Physical and Life Sciences
Energy
Strategic Deterrence
Featured Articles







