Not all bacteria are created equal
 (Download Image)
(Download Image)
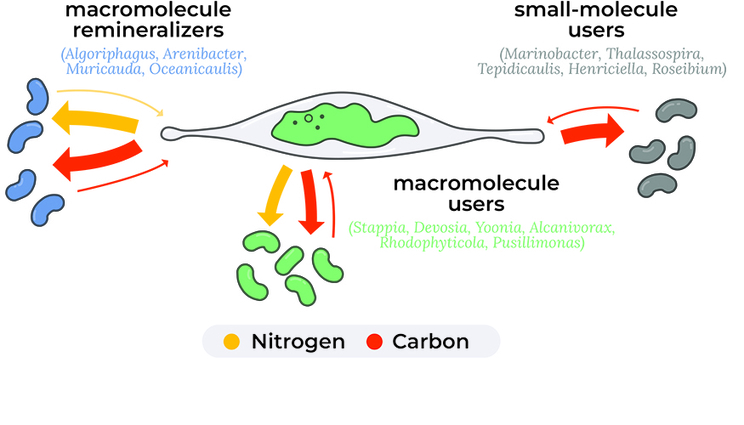
Empirical measurements of carbon and nitrogen exchange between algae and bacteria, using stable isotope tracing, allowed the LLNL team to identify three different bacteria types with distinct ecological roles, providing a conceptual framework to better understand how the algal microbiome plays a role in carbon and nitrogen degradation and recycling.
Lawrence Livermore National Laboratory (LLNL) scientists have found that bacteria can help algae make carbon-neutral biofuels or draw down additional carbon from the atmosphere if the conditions are just right.
The team used LLNL’s nanoSIMS to understand and quantify the role of the algal microbiome in processing algal carbon (C) and nitrogen (N). The research appears in Nature Communications.
“We found that not all bacteria are created equal,” said LLNL scientist and lead author Xavier Mayali. “Some are able to recycle the carbon and nitrogen for the algae to pick up again, and others do not.”
This work is critical to the cost-effectiveness of using algae to make carbon-neutral biofuels or for carbon sequestration from the atmosphere.
Algal-bacterial interactions are fundamental to photosynthetic CO2 fixation in the oceans and other surface waters, including algal bioenergy and bioproduct production ponds. Studies have shown that bacteria can increase algal productivity by providing vitamins, iron-binding compounds and algal growth hormones.
More fundamentally, bacteria can remineralize nutrients, for example, turning organic N to ammonium, a fertilizer that can further increase algal growth. It is generally assumed that algae-associated bacteria grow on algal-derived organic material, making these interactions mutualistic (when both bacteria and algae benefit), but few studies have empirically measured the fraction of bacterial C and N that is derived from photosynthetic microalgae.
“Using the nanoSIMS imaging mass spectrometer enables precise measurements of even low levels of isotope incorporation at the single cell level," said LLNL co-author Peter Weber. Using this approach, the team was able to quantify nutrient transfers between bacteria and algae.
“These microscale exchanges, when integrated over large volumes in outdoor algal cultures and in aquatic ecosystems, have profound consequences for elemental cycling,” LLNL co-author Rhona Stuart said.
One of the goals of this research was to quantify the different rates at which bacteria produce nutrients for algae and develop a rubric for classification.
“We needed to measure net exchanges of C and N between algae and bacteria, identify whether these exchanges are correlated to mutualism or physical association and whether the quantity can be predicted by phylogenetic affiliation or genomic content,” LLNL co-author Ty Samo said.
The team did not find that genomic content could predict biogeochemical activity, suggesting that direct measurements are still needed to understand microbial processing of elements, especially C, in the environment.
“Understanding the cell-specific activity of microbes in their native environment is critical if we are to predict their responses to climate change,” Mayali said. “In the future, we hope to be able to predict microbial C cycling simply from genomic information, but at this time, we don’t know enough, and we have to measure these rates empirically.”
Other LLNL contributors include Jeff Kimbrel, Megan Morris, Kristina Rolison, Courtney Swink and Christina Ramon,as well as researchers from Pacific Northwest National Laboratory.
This work was led by LLNL and funded by DOE’s Office of Science, through the MicroBiospheres Science Focus Area.
Contact
 Anne M. Stark
Anne M. Stark
[email protected]
(925) 422-9799
Related Links
Nature CommunicationsMicroBiospheres Science Focus Area
Tags
Bioscience and BioengineeringNuclear, Chem, and Isotopic S&T
Physical and Life Sciences
Biosciences and Biotechnology
Nuclear and Chemical Sciences
Office of Science
Science
Featured Articles







