Next-generation simulation infrastructure bridges multiscale models
One of the fundamental challenges in computational modeling is to balance the trade-off between the length and time scales that need to be simulated and the corresponding computational costs. Relevant time scales are typically determined by a phenomena of interest, e.g., a protein’s activation or a chemical reaction, whereas length scales are usually chosen based on the amount of detail needed, ranging anywhere from the first-principles representation of elementary particles to the entire cosmos.
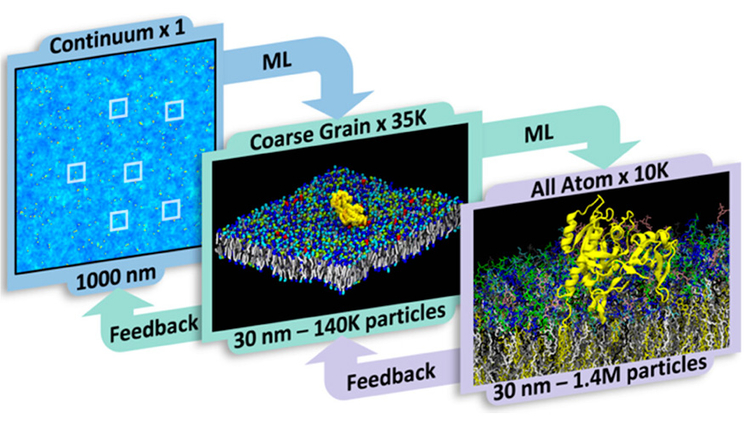
Multiscale modeling and simulation frameworks, such as the Multiscale Machine-Learned Modeling Infrastructure (MuMMI), offer an efficient way to limit unproductive simulations and maximize relevant computation. This method allows MuMMI to combine models at different length and time scales to model the interactions of oncogenic membrane proteins (RAS-RAF). Previously limited to connecting two time and length scales, a multi-disciplinary team of LLNL researchers and collaborators have enhanced MuMMI to include a third scale. This new version of MuMMI represents the first three-scale multiscale approach to explore the RAS/RAF/MAPK signaling pathway that is frequently dysregulated in cancer. When dysregulated, this pathway is a prominent driver in cancer biology.
MuMMI’s original scales are the continuum and coarse grained (CG)—the continuum is the coarsest, providing a zoomed-out overview of the RAS-RAF protein, while the CG provides a more zoomed-in view of protein-lipid interactions. The newest addition to MuMMI—the all-atom (AA) model—is the finest scale and captures specific interactions between lipids and proteins with great detail. AA simulations explicitly represent all atoms and capture details down to hydrogen bonding and vibrations.
A unique capability of the MuMMI multiscale approach is the ability to use the information obtained from a fine-scale simulation to update and improve the coarse-scale simulations while the multiscale campaign is running. Together, these three scales can capture different levels of granularity across time and length scales.
[Helgi I. Ingólfsson, Harsh Bhatia, Fikret Aydin, Tomas Oppelstrup, Cesar A. López, Liam G. Stanton, Timothy S. Carpenter, Sergio Wong, Francesco Di Natale, Xiaohua Zhang, Joseph Y. Moon, Christopher B. Stanley, Joseph R. Chavez, Kien Nguyen, Gautham Dharuman, Violetta Burns, Rebika Shrestha, Debanjan Goswami, Gulcin Gulten, Que N. Van, Arvind Ramanathan, Brian Van Essen, Nicolas W. Hengartner, Andrew G. Stephen, Thomas Turbyville, Peer-Timo Bremer, S. Gnanakaran, James N. Glosli, Felice C. Lightstone, Dwight V. Nissley, and Frederick H. Streitz, Machine Learning-Driven Multiscale Modeling: Bridging the Scales with a Next-Generation Simulation Infrastructure, J. Chem. Theory Comput. (2023), DOI: 10.1021/acs.jctc.2c01018.]
–Physical and Life Sciences Communications Team
Tags
Bioscience and BioengineeringBiosciences and Biotechnology
Physical and Life Sciences
Featured Articles