“Kill switch” design strategies for genetically modified organisms
 (Download Image)
(Download Image)
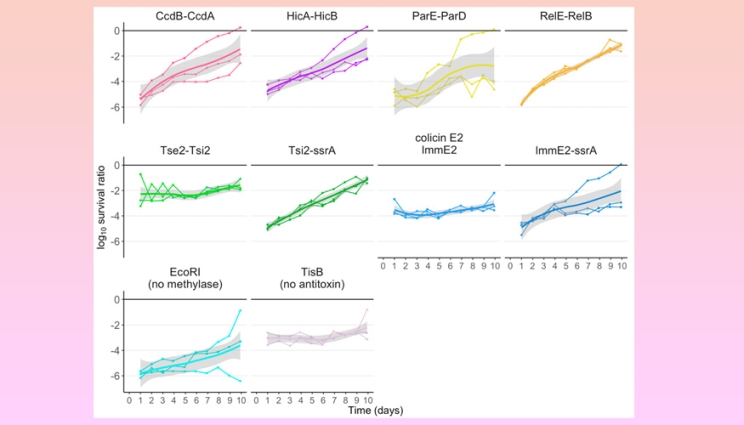
Stability of kill switches during 100 generations of growth. Three independent lineages of each kill switch strain were grown continuously for 10 days.
Synthetic biology promises to sustainably produce commodity chemicals, treat human disease, steward the environment, and sustain agriculture. Each of these tasks exploit genetically modified organisms (GMO), but the unintended release (referred to as “escape”) of GMOs or their recombinant (non-native) DNA could disturb native ecosystems in unpredictable ways. Biocontainment strategies, such as “kill switches,” are therefore needed to limit unforeseen environmental disturbances. Kill-switch circuits prohibit GMO growth by induction of a toxin gene via an environmental signal—i.e., something that turns the kill switch “on,” such as a chemical, pH levels, temperature, or carbon dioxide levels.
To date, the majority of kill switches have been developed in Escherichia coli (E. coli), and tested under ideal laboratory conditions. Although some switch designs have been developed in other host strains—i.e., Pseudomonas putida (bacterium) and Saccharomyces cerevisiae (fungus)—moving kill switch parts that were developed in E. coli into alternative cellular hosts is challenging, resulting in unpredictable changes in kill-switch performance. GMOs developed for environmental applications will ultimately be deployed into uncontrolled environments, where unpredictable inputs and stressors will affect circuit function and failure. As kill-switch optimization strategies are ultimately host dependent, the major modes of escape in each host strain must be determined in order to develop robust kill switches for biocontainment.
Given the lack of containment systems for Pseudomonas fluorescens—a plant-growth-promoting agricultural GMO—a team of BBTD scientists set out to understand how to design kill switches that balance lethality with long-term genetic stability in this important agricultural strain. To do this, the team designed eight different kill switches, each encoded with a lethal toxin and its associated “inactivator” gene that protects the host from lethality. This design provides two knobs that can be turned: one to increase lethality (toxin) and the other to reduce lethality (toxin inactivator).
By studying these kill-switch designs, researchers found that inactivator proteins are powerful determinants of kill-switch functionality and stability. Additionally, an analysis of genetic escape from each kill switch revealed critical determinants of kill-switch stability and important considerations for host-specific circuit design in Pseudomonas fluorescens. Altogether, the results of these in-depth analyses will guide the designs of more stable, highly effective biocontainment systems.
[T.M. Halvorsen, D.P. Ricci, D.M. Park, Y. Jiao, and M.C. Yung, Comparison of Kill Switch Toxins in Plant-Beneficial Pseudomonas fluorescens Reveals Drivers of Lethality, Stability, and Escape, ACS Synthetic Biology (2022), doi: 10.1021/acssynbio.2c00386.]







