Evaluating co-circulating pathogens in COVID-19 samples
 (Download Image)
(Download Image)
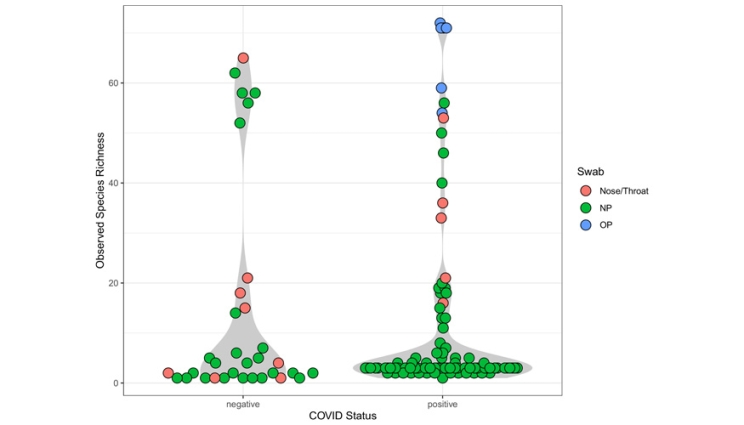
Observed species richness in SARS-CoV-2 positive vs negative samples detected by the LLMDA. Samples with no species detected are not included. The samples were coded by color based on their types: nose/throat swabs are shown in red circles; nasopharyngeal (NP) swabs are shown in green circles; oral pharyngeal (OP) swabs are shown in blue circles. Overall, viral and bacterial taxa were detected from 125 samples (62%), 92 SARS-CoV-2 positive samples and 33 SARS-CoV-2 negative samples. There was no significant difference in the number of species detected in SARS-CoV-2 positive and negative samples.
During the first months of the COVID-19 pandemic, beyond singular infection with SARS-CoV-2, reports of co-infection (or secondary infection) with other respiratory pathogens emerged. Co-infections with SARS-CoV-2 have the potential to affect disease severity and morbidity, however, the potential influence of the nasal microbiome on COVID-19 illness is not well understood.
In collaboration with the California Department of Public Health (CDPH), LLNL researchers analyzed 203 archived COVID clinical samples (101 SARS-CoV-2 positive and 102 SARS-CoV-2 negative samples) to evaluate co-circulating pathogens. In contrast to some earlier studies where samples were collected before February 2020—when other viral pathogens were known to be circulating—most samples in this study were collected after the declaration of a statewide Shelter-in-Place on March 19, 2020 (from February 2020 and July 2020).
The samples were analyzed for the presence of viral, bacterial, and fungal pathogens and non-pathogens using a comprehensive microarray technology, the Lawrence Livermore Microbial Detection Array (LLMDA). The LLMDA has been applied to a variety of human and animal clinical samples to identify pathogens in disease cases and assess the microbiome differences between healthy and diseased samples. Additionally, LLMDA was recently updated to detect SARS-CoV-2, such that a single test run on LLMDA will include SARS-CoV-2 and 12,000 other microbes and viruses, providing more information than a single COVID-19 test.
For their research, the team aimed to evaluate both the burden of co-infections in patients with COVID-19, as well as evidence of differences in the microbiome between SARS-CoV-2 positive vs SARS-CoV-2 negative samples. From the 203 samples, the researchers found sixty-two percent of the samples were positive for one or more viruses and/or bacteria. Beyond SARS-CoV-2, the most prevalent detected pathogens were the bacterial species Streptococcus pyogenes and Streptococcus pneumoniae.
In general, the described study uses a more comprehensive and multiplexed detection platform that enables simultaneous detection of multiple viral, bacterial, and fungal infections, even those not suspected initially.
[J.B. Thissen, M.D. Morrison, N. Mulakken, W.C. Nelson, C. Daum, S. Messenger, D.A. Wadford, C. Jaing, Evaluation of co-circulating pathogens and microbiome from COVID-19 infections, PLOS ONE (2022), doi: 10.1371/journal.pone.0278543.]
Tags
Bioscience and BioengineeringBiosciences and Biotechnology
Physical and Life Sciences
Featured Articles







