Using synthetic biology for chlamydia vaccines
 (Download Image)
(Download Image)
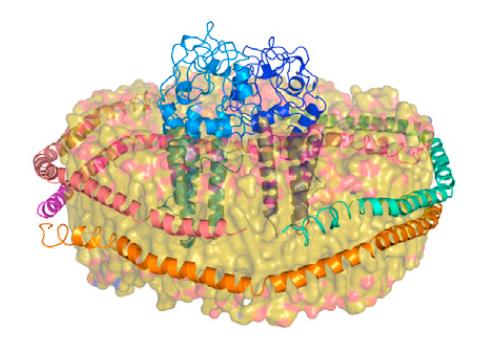
An artist’s rendition of the biological machinery that is involved in the cell-free creation of a major outer membrane protein, supported by Lab-created telodendrimer nanolipoprotein particles.
A multidisciplinary scientific team at Lawrence Livermore National Laboratory (LLNL) has made significant advances in developing a vaccine for chlamydia using synthetic biology, sponsored by a two-year National Institutes of Health (NIH) grant. They detail their work in a recent paper published in the Journal of Biochemistry (JBC): "Cell-Free Production of a Functional Oligomeric Form of a Chlamydia Major Outer Membrane Protein (MOMP) for Vaccine Development." This work was performed in partnership with researchers at University of California, Davis and the vaccine group at Synthetic Genomics, Inc.
Chlamydia is the most common infectious sexually transmitted disease, caused by the gram-negative bacterium Chlamydia trachomatis. The infection is often asymptomatic, and when left untreated, the disease can cause severe chronic health issues such as permanent infertility, pelvic inflammatory disease and blindness. While antibiotic treatments for chlamydia infections exist, recurrences of the disease in the same patient are common and difficult to treat.
"Although antibiotics are used to treat chlamydia, early screening and diagnosis are key to prevent complications associated with long-term, untreated infections," said Wei He, a postdoctoral researcher at the Lab and the primary author of the paper. "It’s important to note that individuals treated with antibiotics are more prone to reinfection, so working toward a vaccine is an essential step in treating this pervasive public health problem."
Vaccines interact with patient immune systems to produce active immunity, which provides protection from disease. They contain antigens — usually purified proteins, like a major outer membrane protein (MOMP) — that are expressed by the pathogen of interest. Upon vaccination, an antigen-specific immune response develops, protecting the patient from real-world infection. An antigen that successfully elicits this immune response is called ‘immunogenic.’
Creating these antigens in the lab, however, can be difficult due to misfolding of the crucial, extremely complex proteins, such as the chlamydia-specific MOMP targeted by this research group. The work discussed in the group’s journal article includes a patent-pending technique unique to Lawrence Livermore (developed in partnership with Synthetic Genomics, Inc.) that has produced high-yield, functional, immunogenic chlamydia MOMP — a breakthrough in MOMP production techniques.
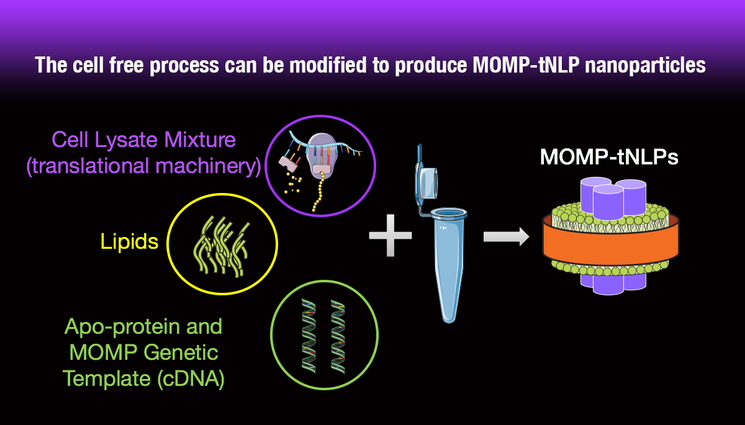
"We’re the first group in the world to use something called telodendrimer nanolipoprotein particles (tNLPs) to produce chlamydia membrane proteins in a cell-free environment," said Matt Coleman, the senior author on the paper.
The only other successful method for creating MOMPs in the lab involves using E. coli as host cells, altering their DNA and using their replication and translation machinery to produce the MOMP, which is then extracted from the E. coli. This method is lengthy and strenuous, and fails at producing a high-yield of correctly folded antigens.
"Our cell-free method lets us take the required tools — the enriched ribosomes and the translational machinery — out of the E. coli. We then add a few more essential components, such as RNA polymerase, to essentially create a 'one-pot' mechanism for protein production outside of a host cell," He said. "Our unique tNLPs are self-assembled within the reaction to produce a kind of scaffolding that supports the MOMP protein in a functional state, even though it doesn’t have a cell environment to support it."
This cell-free method also allows researchers to produce significantly higher quantities of the MOMP because they don’t have to contend with MOMP toxicity to the host E. coli cell — because there is no cell at all. This LLNL-pioneered, single reaction cell-free process removes many of the steps that are required in other methods of producing recombinant proteins associated with nanoparticles and takes less than a day, whereas the other, less effective methods can take up to three days.
"With this paper, we’ve shown that it is possible to take a potentially therapeutic protein that has a very complex structure and recreate it starting with some simple biological components, which is a promising development in this field," added Coleman.
"Chlamydia doesn’t just affect over 131 million people each year worldwide," Coleman said, "but various strains of the bacterium also pose a major problem for animal husbandry. The infection has been particularly devastating for the koalas in Australia. It’s decimated their population."
"Developing this chlamydia vaccine, along with the unique method that we have for making these antigens "on-demand" or as needed for use in vaccines, could be a godsend for the epidemic that’s affecting the koalas, and also obviously has implications for treatment of diseases that affect humans as well — it’s not just chlamydia, there’s a whole host of disease antigens that are notoriously difficult to produce," He added.
The project carries important implications for the development of a vaccine not only for chlamydia, but other bacteria and diseases that also require difficult-to-produce antigens for effective vaccination — including cancer.
Cancer immunotherapy is a burgeoning field aimed at harnessing a patient’s immune system to recognize and attack the unique proteins associated with cancer cells. This could potentially provide another avenue of effective cancer treatment, and the tNLP-supported nanoparticles, coupled with the cell-free method pioneered by the LLNL group (in partnership with Synthetic Genomics, Inc.), could provide a more efficient and effective method of creating antigens for these kinds of cancer vaccines.
The team is now moving forward with trials in mice, and has seen preliminary positive results in producing an immunogenic response that leaves animals with some degree of protection from infection. The next stages of the research involve using the new MOMP production process to engineer the most effective vaccine formula. The project could eventually produce a usable product that could save millions of lives — both koala and human.
The LLNL team also included Nicholas Fischer, Amy Rasley, Craig Blanchette (now at Genentech), Aleksandr Noy, Feliza Bourguet, Angela Evans, Jia Geng and David Homan. This work originally began as a work for others (WFO) endeavor with funding from Synthetic Genomics, and evolved into the two-year, $485,000 grant from NIH. The work in this paper was supported in particular by Luis de la Maza at UC Irvine, Kit Lam and Holland Cheng of UC Davis and Amy Gryshuk of LLNL, who was critical in making the collaboration between LLNL and Synthetic Genomics possible.
Contact
Maren Hunsberger[email protected]
19254226688
Related Links
National Institutes of HealthCell-free Production of a Functional Oligomeric Form of a Chlamydia Major Outer Membrane Protein (MOMP) for Vaccine Development
UC Davis
Synthetic Genomics, Inc.
Tags
Physical and Life SciencesFeatured Articles