Redesign of Cardioid's heartbeat simulation brings code one step closer to clinical use
 (Download Image)
A still from a Cardioid simulation shows the electrical activation of the heart. The colors correspond to the voltage difference across the cardiac cell membrane. Nearby cells signal each other in a propagating electrical wave much like a nerve cell, allowing the heart to coordinate its contraction.
(Download Image)
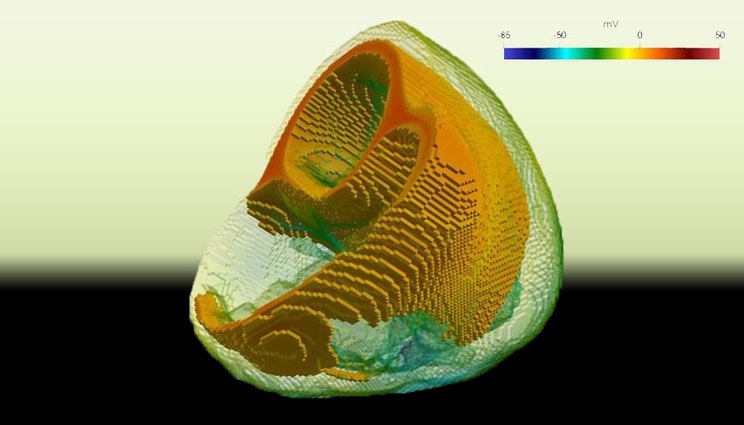
A still from a Cardioid simulation shows the electrical activation of the heart. The colors correspond to the voltage difference across the cardiac cell membrane. Nearby cells signal each other in a propagating electrical wave much like a nerve cell, allowing the heart to coordinate its contraction.
A team of Lawrence Livermore National Laboratory (LLNL) researchers has successfully optimized a code that models the human heartbeat for next-generation, graphics processing unit (GPU)-based supercomputers, with an eye on developing it for virtual drug screening and modeling heart activity in clinical settings.
Cardioid, a suite merging mathematical solvers for electrophysiology, fiber-generation, cardiac mechanics, torso-electrocardiograms (ECGs) and cardiac meshing tools, simulates the electrical current running through the heart tissue, triggering cells to contract like cascading dominoes and causing the heart to beat. It was originally developed by LLNL and IBM for Sequoia, at one time the world’s fastest supercomputer, and was a finalist for the 2012 Gordon Bell Prize, supercomputing’s top honor.
LLNL computational scientist David Richards, one of the original Cardioid developers, recalled that "competing for the Gordon Bell Prize meant customizing the code to take advantage of every bit of performance Sequoia could offer. The resulting code was very fast, but it also was specific to Sequoia and wasn’t portable to other computers."
When LLNL computer scientist Rob Blake came to the Lab in 2016, he was tasked with porting the code over to the heterogenous CPU/GPU systems that would displace Sequoia: Sierra and its unclassified counterpart, Lassen. Blake was one of the founders of a company called Cardiosolv, which used magnetic resonance imaging (MRI) data to model the heart. Cardioid was a good test case for a GPU-based system, Blake said, because the ordinary differential equations (ODEs) required to model the simulation’s 400 million cells — roughly one-fifth of the entire number of cells in the human heart — already were "embarrassingly parallel."
With help from IBM through the institutional Center of Excellence, Blake and his team set out to release Cardioid as open source and to generalize the code for the new supercomputing systems, redesigning the code to be portable as well as fast. He wrote a language called Melody to describe the differential equations used by the solvers to model the heart’s ion pumps and the team ran the code on Sierra and Lassen in 2018, with promising results.
"It was an amazing feat," Blake said. "We were able to test a bunch of new ways to port codes to Sierra and form the other code bases. The code is running quickly on Lassen and we’re getting scientific results out of it that would’ve been impossible if the code was still locked to Sequoia."
Now, with Cardioid working well on GPUs, Blake is heading an internal Laboratory Directed Research and Development (LDRD) project to use Cardioid with machine learning and data from electrocardiograms to determine how the heart is functioning. Using a simple ECG voltage reader, he said, doctors could plug the heart’s electrical activity data into a cellphone to generate a "3D electrical stethoscope." Preliminary results show the approach is working well, he said. "If this technique works, it could be deployed in every operating room in the country," Blake said. "With simulations, I can generate as much virtual data as I want, so I can take that virtual data and use that to train a machine learning algorithm. Once that algorithm has been trained, I could take that over to actual clinical results, and then you develop a machine learning system as a software process that can go through [the Food and Drug Administration]."
The work has other national security implications, as LLNL researchers need the ability to prove pharmaceutical countermeasures to biological agents are safe and won’t negatively affect the heart’s functions, Blake said.
"We want to quickly be able to deliver instantaneous results about whether drugs are going to cause problems from a national security standpoint," Blake said. "The whole idea behind simulation as a science is that we want to simulate things that are too expensive to test. Testing cardiac stuff is really costly in terms of lives of what it takes to get these things working well. If we could simulate that, we could actually bring HPC into medical science."
Blake said he wants to work with the FDA to develop Cardioid for use in a clinical setting and someday virtually screen potential new prescription drugs for their effect on the heart without having to test the drugs on animals. The FDA, for example, is a partner in the Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative, an attempt to reconstruct electrophysiologic activity within a heart cell to help predict negative reactions to a given drug, perhaps replacing animal testing with simulations.
Blake said the "next big thing" in medical research is using simulation the way it’s been used in engineering for years. He believes the next software-only medical device will be released in the next five years, and that Cardioid is "definitely a candidate to run on that track."
"In engineering, we started by building tiny models of bridges and trying to break it, and that’s pretty much all moved over to simulation now," he said. "There’s a real revolution going on in engineering design because of that. Medical is largely missing all of that. To take hard medical problems, simulate them first and study what will happen, and then apply the therapy, that’s going to be the future of medicine."
Other collaborators included project co-developer and LLNL bioscience researcher Xiaohua Zhang, Changhoan Kim of IBM, code developer Jamie Bramwell and HPC systems engineer Phil Regier of LLNL and former LLNL scientist Jean Luc Fattebert, who is now at Oak Ridge National Laboratory.
Contact
 Jeremy Thomas
Jeremy Thomas
[email protected]
(925) 422-5539
Related Links
LLNL ComputationSimulation
Tags
HPC, Simulation, and Data ScienceComputing
HPC Innovation Center
Featured Articles