Playing video games may help researchers find personalized medical treatment for sepsis
A deep learning approach originally designed to teach computers how to play video games better than humans could aid in developing personalized medical treatment for sepsis, a disease that causes about 300,000 deaths per year and for which there is no known cure.
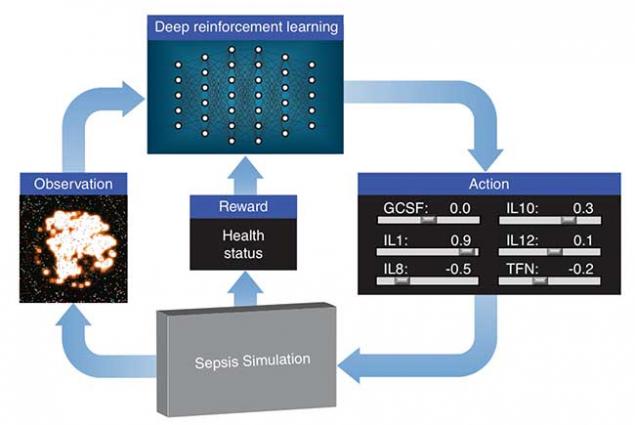
Lawrence Livermore National Laboratory (LLNL), in collaboration with researchers at the University of Vermont, is exploring how deep reinforcement learning can discover therapeutic drug strategies for sepsis by using a simulation of a patient’s innate immune system as a platform for virtual experiments. Deep reinforcement learning is a state-of-the-art machine learning approach originally developed by Google DeepMind to teach a neural network how to play video games, given only pixels as input and the game’s score as a learning signal. The algorithms often exceed human performance, despite not being given any knowledge about the mechanics of the game.
LLNL’s deep learning approach treats the immune system simulation developed by their collaborators as a video game. Using outputs from the simulation, a "score" based on patient health and an optimization algorithm, the neural network learns how to manipulate 12 different cytokine mediators — immune system regulators — to drive the immune response to infection back down to normal levels. The research appears in a paper published by the International Conference on Machine Learning.
"It’s a complex system," said LLNL researcher Dan Faissol, principal investigator of the project. "Previous investigations have thus far been based on manipulating a single mediator/cytokine, generally administered with either a single dose or over a very short course. We believe our approach has great potential because it explores much more complex, out-of-the-box therapeutic strategies that treat each patient differently based on the patient’s measurements over time."
The treatment strategy the researchers propose is adaptive and personalized, improving itself on a feedback loop by continually observing cytokine levels and prescribing drugs specific to the individual patient. Each run of the simulation represents a different patient type and different infection initial conditions.
"The challenge was to keep things clinically relevant," explained LLNL researcher Brenden Petersen, the technical lead for the project. "We had to ensure that all aspects of the simulated problem were relevant in the real world — that the computer wasn’t using any information that wouldn’t really be available in a hospital. So, we only provided the neural network with information that can actually be measured clinically, like cytokine levels and cell counts from a blood draw."
Using the agent-based model with deep reinforcement learning, researchers identified a treatment policy that achieves a 100 percent survival rate for the patients on which it was trained, and a less than 1 percent mortality on 500 randomly selected patients.
"The simulation is mechanistic in nature, which means we can virtually experiment with drugs and drug combinations that haven’t been tested before to see if they might be promising," Faissol said. "The number of possible treatment strategies is huge, especially when considering multi-drug strategies that vary over time. Without using simulation, there’s no way to evaluate all of them. The hard part is discovering a strategy that works for all patient types. Everyone’s infection is different, and everyone’s body is different."
The team’s research has shown that this adaptive approach can lead to novel insights, and the researchers hope to convince others to adopt the approach on sepsis and other diseases.
"Our grand, long-term vision is a ‘closed-loop’ bedside system where measurements from a patient are fed into a decision-support tool, which then administers the correct drugs at the correct doses at the correct times," Petersen said. "Such treatment strategies would first have to be vetted and fine-tuned in wet-lab and animal models, eventually informing real treatments."
Petersen said most of the hardware to execute such a closed-loop system already exists, as with simpler systems like insulin pumps that constantly monitor the blood and administer insulin at the right time.
The Lab’s deep reinforcement learning approach has yet to be tested in the real world, but based on the success using the simulation, the National Institutes of Health awarded LLNL and the University of Vermont researchers with a five-year grant to continue the work, primarily on sepsis but also on cancer.
"This is an exciting project," said Gary An, a critical care physician at the University of Vermont and computational scientist who developed the original version of the sepsis simulation. "This is an incredibly novel project that brings together three cutting-edge areas of computational research: high-resolution multi-scale simulations of biological processes, extension of deep reinforcement learning to biomedical research and the use of high-performance computing to bring it all together."
LLNL’s director of Bioengineering Shankar Sundaram described the approach as "an illustrative example of the Lab contributing to the development of a potential therapeutic solution to a complex health problem critical to our biosecurity mission, applying and advancing our state-of-the-art capabilities in scientific machine learning and targeting improved causal, mechanistic understanding."
LLNL researchers also have initiated a collaboration with Moffitt Cancer Center in Florida to see if a similar approach could learn effective drug therapy strategies using a simulation of cancer. Moffitt released a video game version of their simulation called "Cancer Crusade" that runs on mobile phones.
"One strategy is to crowdsource the learning by analyzing treatments recorded from the top scoring players around the world," Petersen said. "We applied our deep learning approach and want to see how our computed treatments stack up against the top players — a ‘man vs. machine’ showdown."
The sepsis project also has led to a new effort at LLNL researching adaptive and autonomous cyberdefense strategies using simulation and deep reinforcement learning.
Other team members include LLNL researchers Claudio Santiago and Tom Desautels, LLNL summer interns Jiachen Yang and Clayton Thorrez and Chase Cockrell from the University of Vermont. Funding for the project comes from the Laboratory Directed Research and Development program.
Contact
 Jeremy Thomas
Jeremy Thomas
[email protected]
(925) 422-5539
Related Links
Data ScienceData Analytics
International Conference on Machine Learning
Paper: International Conference on Machine Learning
Tags
HPC, Simulation, and Data ScienceComputing
Engineering
Featured Articles