LLNL’s Brase discusses advances by ATOM in accelerating drug discovery pipeline
The private-public Accelerating Therapeutic Opportunities in Medicine (ATOM) consortium is showing “significant” progress in demonstrating that high performance computing (HPC) and machine learning tools can speed up the drug discovery process, ATOM co-lead Jim Brase said at a recent webinar.
Brase, Lawrence Livermore National Laboratory’s (LLNL) deputy associate director for Data Science, leads the Lab’s efforts in HPC for life science and biosecurity. He said in its nearly five years of existence, ATOM has accelerated generative design and molecular optimization, promising “real and important applications” to future therapeutics for cancer and other infectious diseases.
“We’ve successfully demonstrated that we can scale up and simultaneously optimize (drug molecules) over many parameters and over relatively large molecular populations,” Brase said. “And we think that these systems have the potential to guide and optimize experimental data collection — but there’s still a lot of work to do. (Active learning) is where we’re really focusing on in the future, because I think that’s going to be incredibly useful and very exciting to work on.”
Founded in 2017 by LLNL, the Frederick National Laboratory for Cancer Research, the University of California, San Francisco (UCSF) and pharmaceutical company GlaxoSmithKline, the ATOM consortium currently boasts more than a dozen member organizations, including national laboratories, private industry and universities.
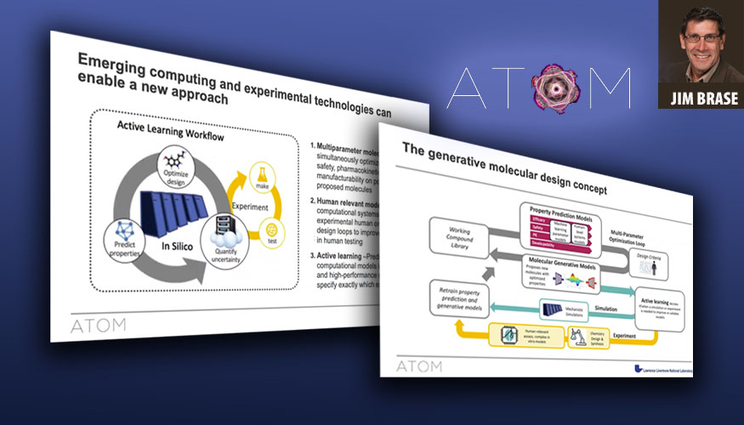
During the webinar, Brase discussed ATOM’s approach to multiparameter molecular design — a machine learning-backed generative loop that predicts properties of proposed drug molecules, screens them virtually for safety, pharmacokinetics, manufacturability and efficacy, optimizes the designs and uses computational models and experimental feedback from the synthesized compounds to improve the models. The work is performed on large-scale parallelized supercomputers at LLNL and elsewhere.
“There’s still a strong experimental component in the approach, but we’re trying to use computation to do the critical experiments that are really necessary to accelerate and make the whole process better,” Brase explained.
Brase pointed to three projects funded by ATOM that have proven the effectiveness of the generative design approach: a demonstration project to improve the selectivity of compounds for inhibiting aurora kinases; an “engineering test” with biotech company Neurocrine Biosciences to optimize histamine antagonist designs (which reduced the timeline for optimizing novel drugs to only a couple of months); and a third project with the UCSF Cancer Center to improve the pharmacokinetic properties of approved cancer drugs while maintaining safety and efficacy. While the latter project is in its early stages, Brase said, the initial results are promising.
“This is the first project where we have a really interesting drug target and are actually trying to produce novel molecules for a specific target,” Brase said. “We’re moving the population and getting substantial numbers of molecules with over 100x selectivity (over existing drugs).”
In addition to providing an overview of ATOM and its accomplishments to date, Brase also presented the technology used in the drug design loop, including the ATOM Modeling PipeLine (AMPL) framework, which he said has proven useful for integrating multiple data sets and training large populations of models. The framework also has shown promise for introducing students to machine learning for drug design through ATOM-related engagements with Purdue University and Butler University. ATOM has made AMPL available as open source, as is the Livermore Big Artificial Neural Network Toolkit (LBANN) used to train the ML models. ATOM’s Generative Molecular Design software also is in the process of becoming open source, Brase said.
“One of goals of ATOM is to make these tools widely available to get people started in this (area) and increase the capability of lots of different groups out there to do some level of molecular design,” Brase said.
In training the machine learning models on molecular data, ATOM researchers have typically relied on a junction tree autoencoder, but the approach limits the speed and the size of molecules that can be reconstructed, Brase explained. ATOM is pursuing a new training structure with Wasserstein autoencoders — a character-based language model that can classify valid molecules at each stage of the training process.
The advantage of Wasserstein encoders is that they can scale up to very large training sets, use larger molecular structures and are less computationally intensive. LLNL scientists used the technique in their 2020 paper on drug discovery for COVID-19 that was a finalist for the ACM Gordon Bell Prize for High Performance Computing-Based COVID-19 Research. The paper showed the encoder could be used to scale up to 17,000 GPUs in parallel on LLNL’s Sierra supercomputer, training on 1.6 billion small-molecule compounds and one million additional promising compounds for COVID-19 while reducing the model training time from one day to just 23 minutes.
Looking ahead, Brase said ATOM hopes to increase parallelization and add more biological features to its machine learning models to improve predictions and increase the speed of transitioning optimized molecules to humans and the clinical environment. A project at UCSF that focused on molecules crossing the blood-brain barrier incorporated biological features and resulted in a “significant improvement” in performance, Brase explained.
“This is turning from a marginally useful model to a clearly useful model with only a small amount of biological test information,” Brase said.
As it continues to add more members, ATOM is transitioning into a 501c3 nonprofit organization — the ATOM Research Alliance — which will give the organization more flexibility to expand, Brase said.
To find out more about ATOM, or how your organization can join the alliance, visit the web.
Contact
 Jeremy Thomas
Jeremy Thomas
[email protected]
(925) 422-5539
Related Links
ATOM scienceTags
ScienceFeatured Articles