LLNL team models COVID-19 disease progression and identifies risk factors
 (Download Image)
(Download Image)
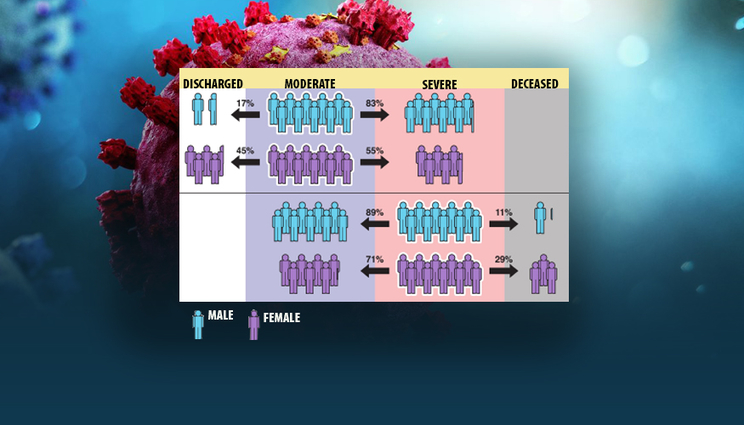
Being male is a known risk factor for adverse outcomes in hospitalized COVID-19 patients. However, new analysis reveals that when modeling the entire disease trajectory, the degree to which being male is a risk factor depends on the underlying disease severity of the patient. Foreground image credit: LLNL Principal Investigator Priyadip Ray; Background image credit: Adobe Stock images.
A Lawrence Livermore National Laboratory (LLNL) team has developed a comprehensive dynamic model of COVID-19 disease progression in hospitalized patients, finding that risk factors for complications from the disease are dependent on the patient’s disease state.
Using a machine learning algorithm on a dataset of electronic health records (EHRs) from more than 1,300 hospitalized COVID-19 patients with ProMedica — the largest health care system in northwestern Ohio and southeastern Michigan — the team classified patients into “moderate” or “severe” states and tracked disease trajectory as patients moved through different risk states during hospitalization.
Accounting for disease severity — in contrast to previous scientific literature examining only static risk factors — the method allowed the team to identify, as the disease progressed, when certain variables such as age and race, and comorbidities including diabetes and hypertension, led to more severe outcomes.
The model allowed the team, which included co-authors from the University of Toledo, to demonstrate for the first time that links between some factors and more adverse outcomes from COVID-19 can depend on the patient’s “current” condition. Most significantly, while male patients were found to be more likely than female patients to have serious complications or die from COVID-19, when starting from the “severe” disease state, women were more likely than men to die of the disease. The results were published Feb. 7 in the Journal of the American Medical Informatics Association.
“It’s well known in the community that men are at a higher risk than women for eventual death from COVID, and that’s true — but certain counterintuitive behavior emerges once you break up the patient trajectory into disease states,” said LLNL principal investigator Priyadip Ray. “From the moderate disease state, men are more likely to transition to a more severe disease state. However, if you are in the severe disease state, surprisingly, women are more likely to die than men. This disease-state perspective has not been shown before and indicates that where you are in your disease also determines your risk factors.”
By modeling the entire trajectory of hospitalized COVID-19 patients, the team showed “statistically significant differences” in the relative risk of disease progression, which they concluded should be taken into consideration when performing risk assessment among patients in hospitals.
“The vast majority of studies on COVID-19 risk factors ignore the temporal progression of the disease in their analysis,” said LLNL co-author Braden Soper. “Our study provides a unique modeling-based approach to understanding how patient demographics and medical comorbidities can present different risk profiles depending on the underlying disease state. Such information is potentially more actionable throughout the course of care, possibly leading to better patient outcomes.”
Soper added that disease state-dependent risk assessment also can apply to many other acute and chronic diseases beyond COVID-19, which have thus far largely been assessed only with static data and modeling techniques.
Since EHRs typically suffer from irregularly sampled and/or missing data, the team used a statistical model known as a covariate-dependent, continuous-time hidden Markov model (HMM), known to handle such data well.
The models showed that, while being male, Black or having a medical comorbidity were all associated with an increased risk of progressing from moderate to severe disease states, the same factors resulted in a decreased risk of transitioning from a severe state to death. Researchers attributed the counterintuitive results to the existing prevalence of static models for risk stratification.
“A fixed-time (static) model is susceptible to immortal time bias, as periods of follow-up may be incorrectly assigned to a particular disease state,” Ray said. “An HMM is less susceptible to such biases, as it can infer the disease state throughout the patient trajectory.”
Among the other findings: body mass index (BMI) alone was not linked to an increased risk of disease progression, while old age was associated with an increased risk in progressing from moderate to severe and from severe to deceased states, the researchers reported.
The team validated the inferred latent disease states with National Institutes of Health-established guidelines and the Epic Deterioration Index risk metric.
Tests on a budget
The LLNL/University of Toledo/ProMedica team’s work on dynamic models follows an earlier paper the team published in Scientific Reports, where they examined static risk factors for patients who go on to develop severe complications after testing positive for COVID-19.
The team used an interpretability tool to find out which lab tests were most predictive for hospitalization or poor outcomes, identifying which tests should be collected in the case of budget constraints that could give clinicians nearly the same predictions for adverse outcomes as collecting all possible data.
“We tried to look at this problem in a different way,” Ray said. “We asked, ‘what if you have a budget constraint? What are the biomarkers that you can collect that will give you a good indication of how likely it is that this patient will need to be ventilated or likely to die due to the disease?’
“The interesting thing is that beyond a certain point, collecting more labs will not necessarily give better predictive performance. Can you select a small set of labs and markers that is indicative of risk?” Ray continued. “The answer we found was yes.”
To make that determination, the team created a cost structure, grouping types of lab tests and biomarkers with associated costs, from free information (demographics and comorbidities) and low-cost tests such as blood pressure and pulse oximetry, to more expensive lab results — such as liver function and inflammation.
The team then used a machine learning method known to work well for healthcare datasets with missing and/or skewed data to find correlations between patient's features and their risks for death or ventilation from COVID-19 and determine the most predictive set of features. Using the method, they found it was possible to achieve a 43 percent reduction in lab costs with only a 3 percent reduction in performance in predicting the likely need for ventilation from the disease.
“We found that it is possible to achieve a significant reduction in cost at the expense of a small reduction in predictive performance,” said co-author Sam Nguyen, who led the data extraction and modeling effort from the LLNL side. “In responding to the pandemic, if one is limited by their testing (vitals, labs, etc.) resources, one could actually do quite well at COVID-19 risk prediction with a limited number of inputs, if one knows the most cost-efficient tests to carry out.”
The findings could be useful where certain lab work is unavailable or prohibitively expensive — of particular importance during the COVID-19 pandemic, where many patients and clinicians need to make quick decisions, researchers said.
Co-authors on both papers included LLNL scientists Jose Cadena and Ryan Chan; Paul Kiszka, Lucas Womack and Mark Work of ProMedica; and Joan Duggan, Steven Haller, Jennifer Hanrahan, David Kennedy and Deepa Mukundan of the University of Toledo.
LLNL’s Laboratory Directed Research and Development program funded both efforts.
Contact
 Jeremy Thomas
Jeremy Thomas
[email protected]
(925) 422-5539
Related Links
Journal of the American Medical Informatics AssociationTags
ASCHPC, Simulation, and Data Science
Computing
Engineering
Science
Featured Articles







