Lawrence Livermore researchers release 3D protein structure predictions for the novel coronavirus
 (Download Image)
(Download Image)
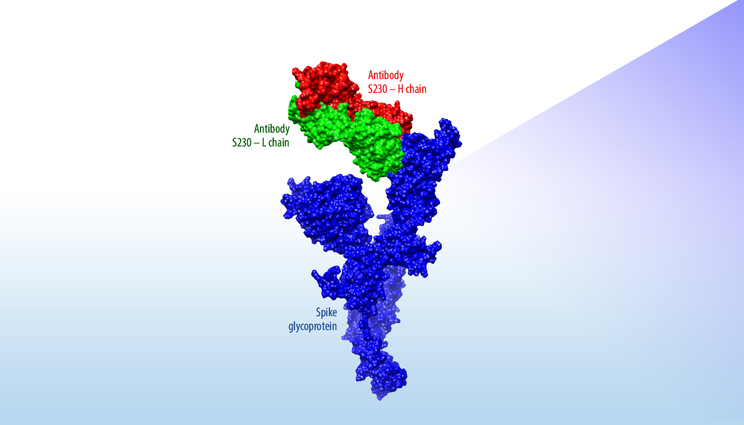
This figure shows how S230, an antibody known to have neutralizing activity against the Severe Acute Respiratory Syndrome coronavirus, is predicted to interact with an initial model of a surface protein on the novel coronavirus. This computational estimate can quickly provide researchers with structural insights without waiting for time-consuming X-ray crystallography images of the actual protein.
As global concern continues to rise about a novel coronavirus spreading from China, a team of Lawrence Livermore National Laboratory (LLNL) researchers has developed a preliminary set of predictive 3D protein structures of the virus to aid research efforts to combat the disease.
The team’s predicted 3D models, developed over the past week using a previously peer-reviewed modeling process, are based on the genomic sequence of the novel coronavirus and the known structure of a protein found in the virus that causes Severe Acute Respiratory Syndrome (SARS), also a coronavirus that closely resembles the new virus.
“A major part of the value of these new structural models is that they present the predicted protein in complex with SARS-neutralizing antibodies,” said Adam Zemla, an LLNL structural biologist and mathematician. “This can be thought of as the first step for the global research community to identify and model how therapeutic antibodies can be designed to fight the novel coronavirus.”
Lab researchers are designing a diversity of protein models because the new coronavirus protein structure is not yet known, according to Daniel Faissol, a data scientist in the Lab’s Computational Engineering Division.
The gold standard for obtaining protein structures for viruses is X-ray crystallography, but the entire process to learn the 3D structure of proteins can require weeks to months.
“We are making our initial protein structures available to the broader research community in the hopes of accelerating the development of disease countermeasures globally, because we hope to see a rapid response and because the situation is evolving so quickly. As we obtain more information about the virus, our intention is to update our models and provide follow-up data releases,” Faissol said.
To date, the Lab researchers have developed seven 3D, predictive models of coronavirus proteins where therapeutics could be targeted using three different antibodies.
The models were developed at LLNL using published methods on structure modeling and structure variability analysis systems and structure alignment software. The most current 3D models can be obtained by contacting the biosecurity [at] listserv.llnl.gov (LLNL Biosecurity Center).
The team’s work in support of the novel coronavirus research is part of an ongoing research effort with the Department of Defense and others working toward accelerating the design of vaccines and therapeutics for various diseases.
As a next step, the team plans to use the preliminary protein models as part of a novel approach for accelerating countermeasure design, using a new LLNL-developed system that combines machine learning, biological experiments and simulation on high performance computing. In this process, the starting point is the estimated protein structures that they recently released.
Ultimately, the system may help identify new and/or improved candidates for countermeasure development. “Being able to estimate these structures rapidly is a key enabler for rapid computational design,” said Thomas Desautels, an LLNL data scientist.
The coronavirus work leverages science and technology capabilities developed under LLNL’s internal Laboratory Directed Research and Development program, which supports cutting-edge, high-risk research.
In addition to Zemla, Desautels and Faissol, other team members are Edmond Lau, a computational chemist, and Magdalena Franco, a biomedical researcher.
Contact
 Stephen Wampler
Stephen Wampler
[email protected]
(925) 423-3107
Related Links
LLNL Biosecurity CenterLLNL Press Release "Public-private consortium aims to cut preclinical cancer drug discovery from six years to just one"
Tags
Global SecurityPhysical and Life Sciences
Featured Articles







