Building a secure biosystem to protect microorganisms
 (Download Image)
(Download Image)
The LLNL Biosecurity Science Focus Area seeks to design multilayered containment strategies to safeguard the deployment of engineered microbes in the rhizosphere.
To reduce the risk of unintended ecological consequences from environmentally deployed, genetically engineered microorganisms (GEMS), Lawrence Livermore National Laboratory (LLNL) scientists and collaborators are developing built-in “security mechanisms” that ensure they function where and when needed.
The team hopes to stabilize GEMs to prevent the transfer of potentially “invasive traits” to neighboring native microorganisms and to control the niche-specific function of GEMs for safer and more effective environmental applications.
LLNL and partners received $9 million from the Department of Energy’s Office of Science, Biological and Environmental Research (BER) program for starting a new Science Focus Area (SFA) as part of BER’s Secure Biosystems Design Program.
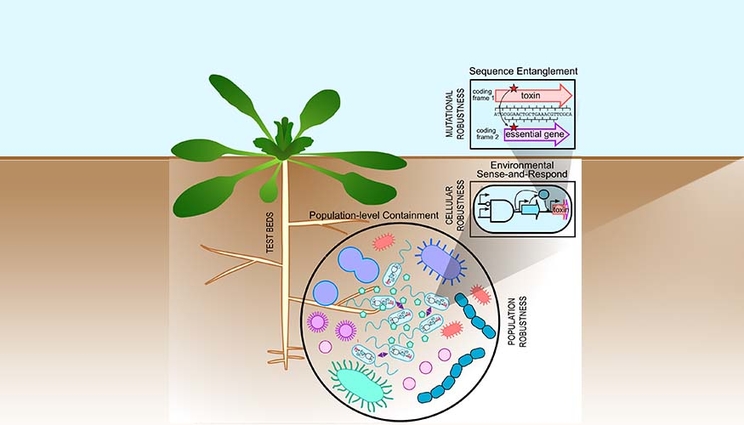
The multilayered containment strategies will help safeguard the deployment of engineered microbes in the rhizosphere (the narrow region of soil that is directly influenced by root secretions and associated soil microorganisms known as the root microbiome).
“Our designs will provide a knowledge base necessary to enable safer use of GEMs in such distinct applications as plant probiotics, carbon sequestration, critical metal recovery, nuclear-activity detection and the production of biomaterials,” said Yongqin Jiao, LLNL environmental microbiologist and lead principal investigator.
The need for biocontainment is especially relevant for environmental biotechnologies for sustainable development of bioenergy crops and carbon sequestration. The microbiota colonizing the rhizosphere of plant roots, especially the plant-benefiting microorganisms (PBMs), contribute to plant growth and modulate soil carbon input, release and storage. Genetic engineering approaches have been used for enhancing the beneficial traits of PBMs, such as nutrient acquisition and drought resistance,
“Because these operations could involve deliberate, large-scale field applications, there is a strong need to understand and prevent potential negative ecological consequences related to GEM persistence in soils and the rhizosphere,” Jiao said.
“Fitness is an important consideration in the development of biocontainment mechanisms,” said LLNL’s Dan Park, a co-prinipal investigator. “For robust environmental function, we need simple but effective biocontainment designs with minimal metabolic burden or background toxicity,”
Layered containment strategies at the sequence, cellular and population levels are expected to increase the overall system robustness to environmental fluctuations.
“We believe that population coordination among engineered microbes also can be exploited as novel design strategies for containment,” said LLNL’s Mimi Yung, a co-principal investigator. “Leveraging native bacterial signaling systems to control collective behaviors has the potential to enhance system responsiveness both temporally and spatially.” These behaviors are especially beneficial in heterogeneous rhizosphere environments where large chemical gradients are present at a small physical scale."
Leveraging LLNL’s high performance computing and high-throughput gene-editing capabilities, the team will advance the synthetic gene entanglement strategy for containment. Two genes are encoded within the same sequence space to protect engineered functions against mutational deactivation or to mitigate the horizontal transfer of potentially invasive traits.
“It is an exciting opportunity to apply machine learning to novel sequence designs that not only improve gene fitness and function but also their evolutionary stability,” said LLNL’s Jonathan Allen, a co-principal investigator. “This work leverages our existing machine learning capabilities established under the Director’s Predictive Biology Initiative through programs like ATOM and positions us to contribute to this new research area of gene design.”
The team will leverage the LLNL Soil Microbiome SFA led by Jennifer Pett-Ridge and the rich experience in soil microbial ecology to test the effectiveness and evaluate the ecological consequences of containment designs in soil and rhizosphere environments.
LLNL scientists Jeffrey Kimbrel, Ali Navid, Adam Zemla and Matthew Coleman also contributed to this work as well as researchers from Columbia University, the University of Maryland, the University of Minnesota and the University of California, Berkeley.
Contact
 Anne M. Stark
Anne M. Stark
[email protected]
(925) 422-9799
Related Links
Biological and Environmental Research programTags
Bioscience and BioengineeringBiosciences and Biotechnology
Physical and Life Sciences
Featured Articles







